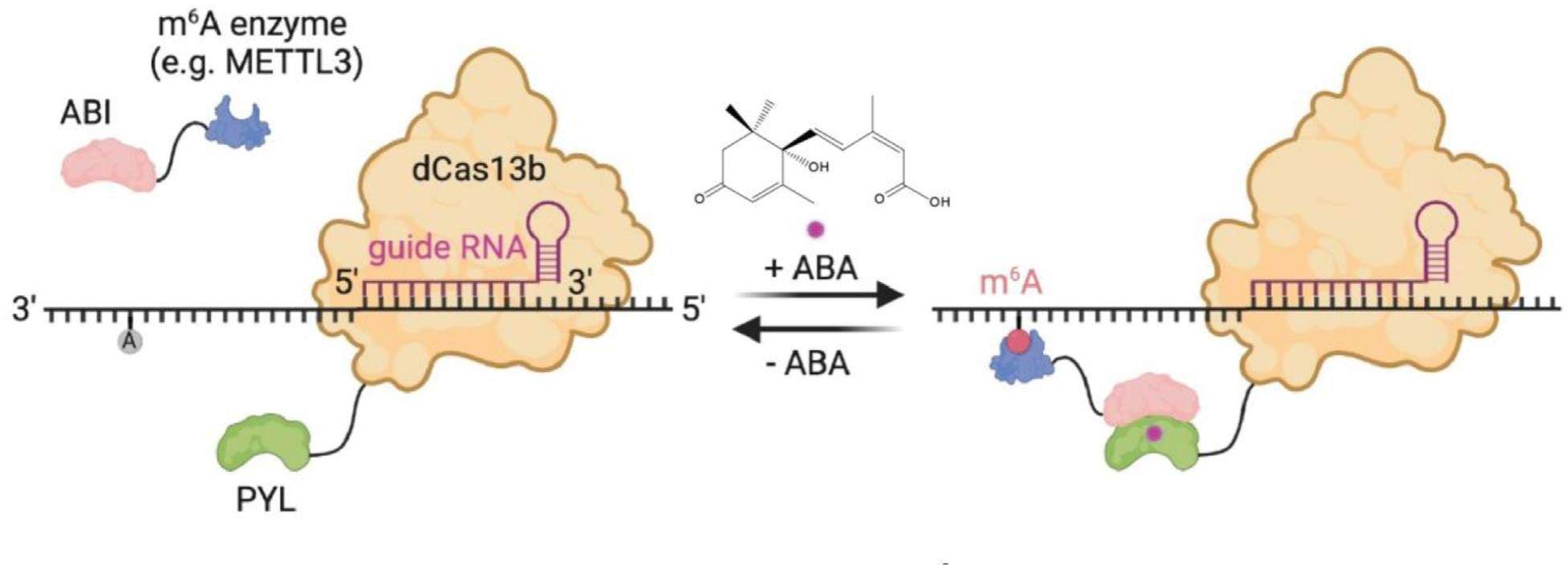
On demand CRISPR-mediated RNA N6-methyladenosine editing


There are over 170 types of chemical modifications discovered across all the RNA categories in living organisms, which constitute the epitranscriptome. Emerging as an additional mechanism of post-transcriptional regulation of genetic information, these RNA modifications and the epitranscriptome have been tightly associated with the RNA fate and various human diseases. Among these distinct RNA modifications, m6A is the most prevalent and involved in coordinating many aspects of biological activities. It has been known that the deposition and removal of the m6A modification are dynamic and reversible processes in cells and regulated by different groups of enzymes and proteins, including the m6A writers, erasers and readers. The m6A writers include the methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14) and Wilms’ tumor 1-associating protein (WTAP), which are responsible for adding the methyl group to N6 position on adenosine to generate the m6A modification, and the fat-mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) have been identified as the enzymes to remove it. The YTH family and heterogenous nuclear ribonucleoproteins (HNRNPs) as well as the eukaryotic initiation factor (eIF) and insulin like growth factor 2 (IGF2) are discovered to be the endogenous proteins to recognize the m6A modification and mediate its biological functions. Dissecting the precise roles of m6A in biological or disease processes has been complicated by the fact that the m6A's function is position, transcript, cell and tissue contextdependent. Conventional genetic methods manipulating m6A writers and erasers not only lead to the global change of m6A levels across the whole transcriptome but also lack temporal controls. Without proper research tools, the relationship between m6A and the phenotypic outcomes within particular RNA and cellular contexts remains elusive.