CTpredX: Enhancing missense variant pathogenicity prediction in childhood cancer predisposition genes


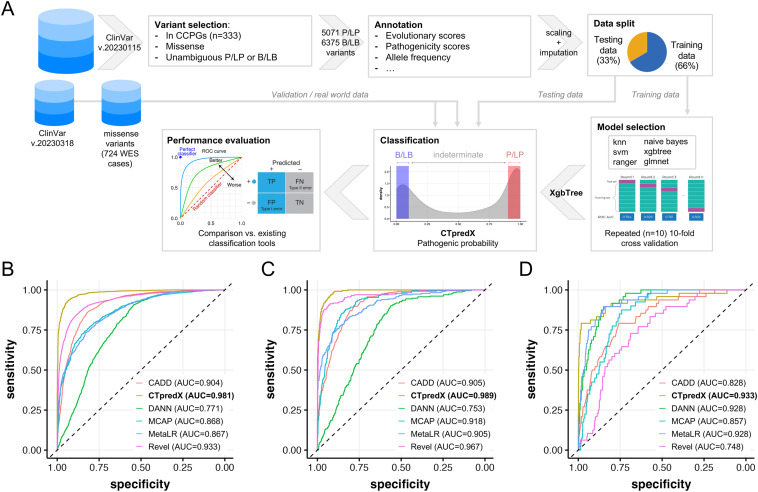
Effective clinical genome interpretation relies on accurately distinguishing between benign and pathogenic rare variants. Current machine learning-based variant prioritization tools are trained on genome-wide data and often overlook key parameters defining gene–disease relationships. Genes that cause a specific disease or a group of related diseases are likely involved in common biological processes. We hypothesize that these genes will share more features not captured by existing genome-wide tools. Disease-specific variant classifiers have been shown to outperform genome-wide tools when specifically applied to inherited cardiac diseases, inherited retinal diseases, or primary immunodeficiencies.1 However, no tool or predictor has been specifically designed for pathogenicity prediction of missense variants in childhood cancer predisposing genes (CCPGs).