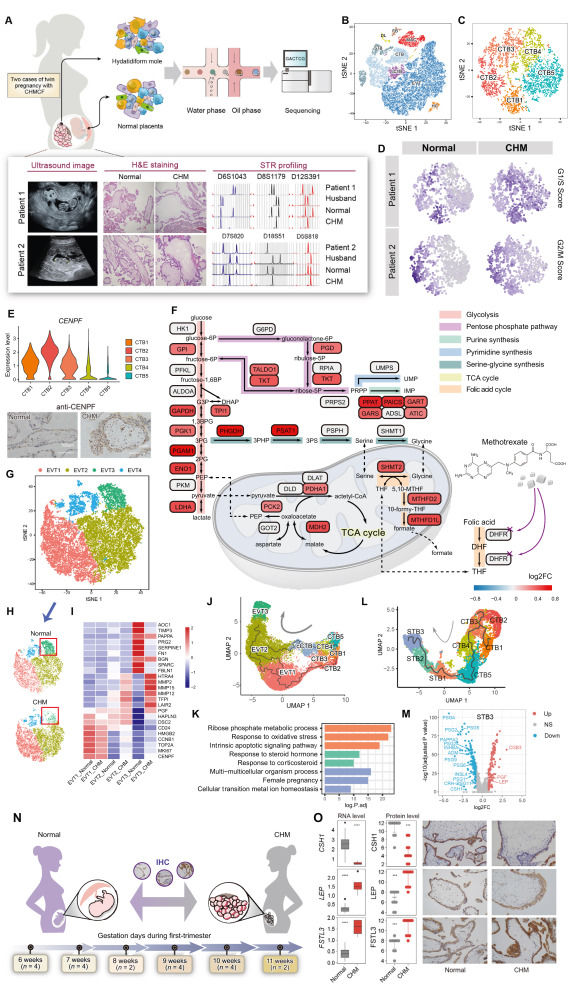
Dissecting trophoblastic heterogeneity in abnormal pregnancy: Insights from comparative analysis of twin-pregnancy with hydatidiform mole and coexisting live fetus


Complete hydatidiform mole (CHM), a typical gestational trophoblastic disease (GTD), is a consequence of abnormal fertilization.1 Historically, trophoblastic cells have been categorized based on morphological and pathological characteristics, with functional classification yet to be well-established. Genetic abnormalities in these cells have been primarily studied using standard techniques such as short tandem repeats (STR) genotyping or microarray comparative genomic hybridization (aCGH).2 Previous studies using single-cell RNA sequencing (scRNA-seq) have uncovered the transcriptomic map of normal human placental cells.3, 4, 5 However, the key genes and transcriptional characteristics of trophoblast differentiation under pathological conditions remain largely unexplored due to genetic complexity and individual heterogeneity. We performed scRNA-seq on villus tissues freshly collected from two Chinese twin-pregnancies with CHM and coexisting live fetus (CHMCF) to compare the distinct transcriptional profiles. In both patients, STR analysis revealed that the molar tissues contained only paternal STR loci, confirming their identification as androgenetic monospermic CHM (Fig. 1A). In a twin pregnancy model of CHMCF, both the CHM and live fetus have the same gestational age (i.e., the timing of trophoblast differentiation) and are exposed to the same uterine environment. This theoretically identical developmental context provides optimal conditions for a comparative study.