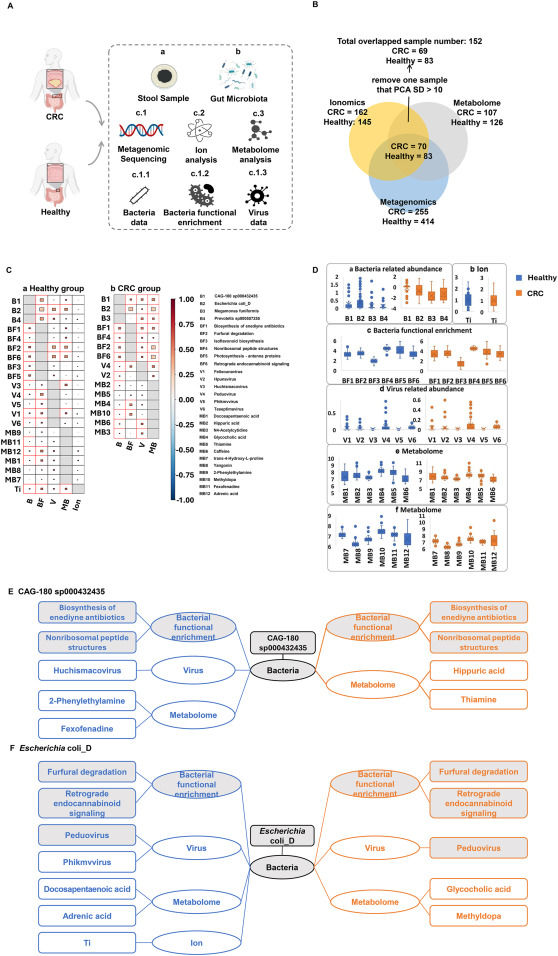
Gut microbiome–metabolome–ionome network spectrum mapping of colorectal cancer


Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, and its occurrence and development are associated with a variety of factors,1 among which the roles of gut microbiota, metabolites, and ions are increasingly valued. However, the interaction relationship between intestine and intestinal microecology is complex, and it is difficult to explain the causal relationship between intestinal microbial interactions with current analytical methods. Structural equation modeling is a statistical modeling method for representing, estimating, and displaying a relationship network between variables.2,3 This method integrates factor analysis and path analysis to examine the complex relationships among multiple variables, thereby systematically elucidating the causal mechanisms underlying clinical phenomena. In this study, intestinal microbiome, metabolome, and ionome were detected in stool samples from healthy individuals and CRC patients, and multi-omics big data was used to map intestinal microecological composition. The structural equation model was used to construct the intestinal microbiome–metabolome–ionome network, and comprehensively illustrate the causal relationships among the complex and diverse influencing factors of CRC, providing a new perspective for revealing the pathogenesis of CRC.