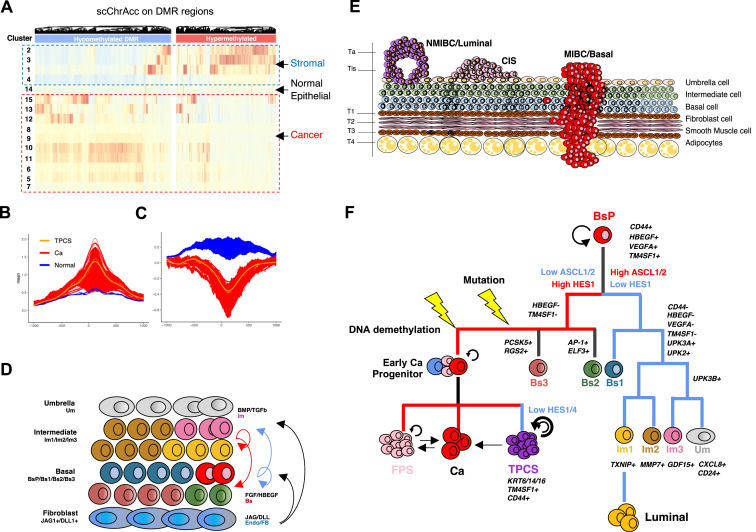
Elucidating the pathogenesis of bladder cancer through single-cell chromatin accessibility and DNA methylation analysis


Previous studies have sought to classify bladder cancer (BLCA) into different molecular subtypes to understand its pathogenic pathways and uncover specific treatments.1 These subtypes, often based on genetic, transcriptomic, or proteomic profiles, aim to stratify patients for precision medicine and improve therapeutic outcomes. Despite these efforts, such classifications have rarely been applied in clinical practice due to challenges in standardization, reproducibility, and limited translational studies validating their utility.1 The treatment of BLCA predominantly relies on surgery, often combined with chemotherapy, immunotherapy, targeted therapy, or antibody-drug conjugates. Radical cystectomy remains the cornerstone for muscle-invasive bladder cancer (MIBC), while transurethral resection and intravesical therapy are common for non-muscle-invasive bladder cancer (NMIBC).2 However, the choice of its treatment modality still depends specifically on whether the disease is NMIBC or MIBC, rather than on the various molecular subtype classifications.3 Bridging the gap between molecular research and clinical application remains a significant challenge, highlighting the need for robust biomarker validation and the development of treatment algorithms that incorporate these subtypes to better guide personalized therapy.