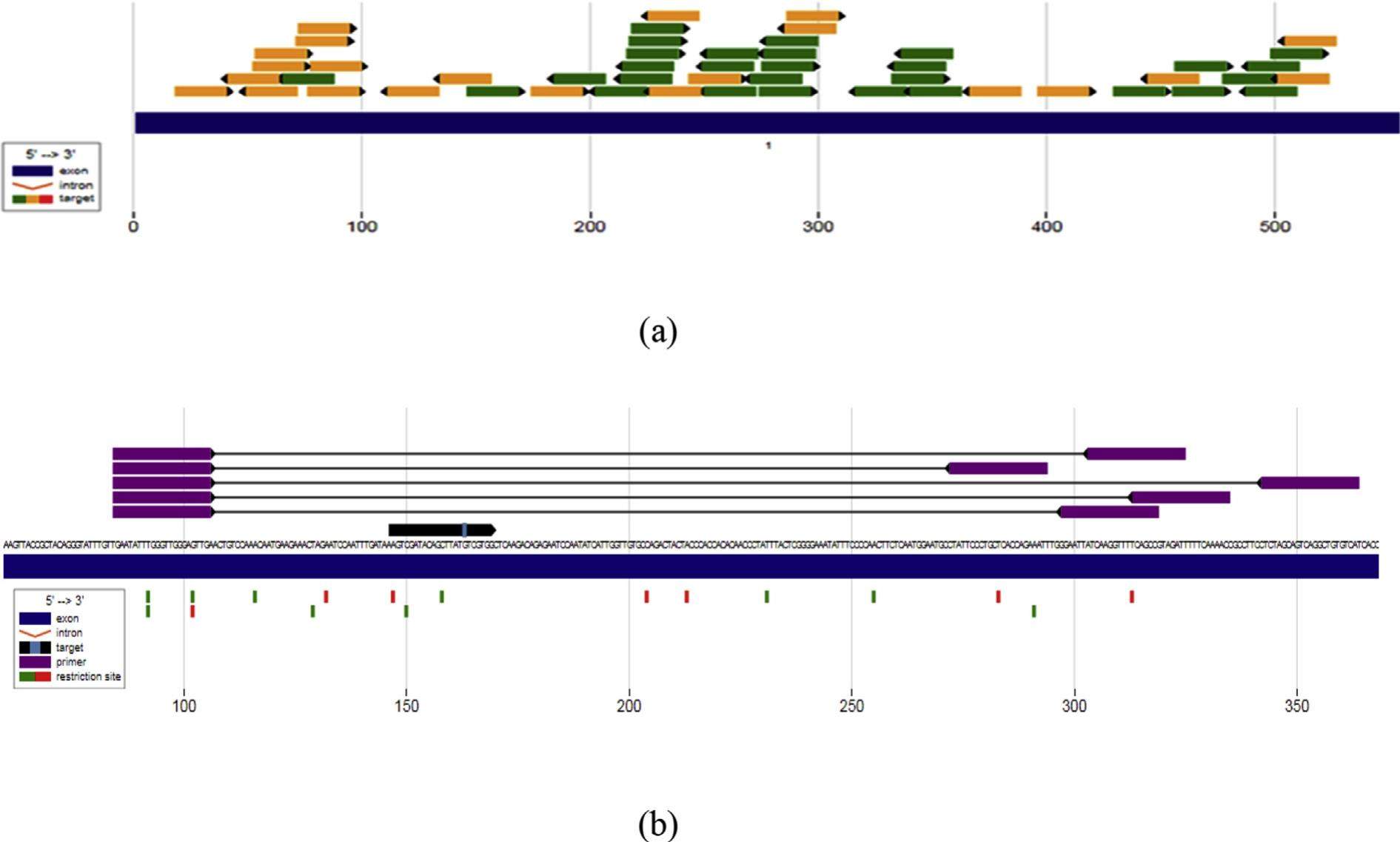
In silico sgRNA tool design for CRISPR control of quorum sensing in Acinetobacter species


CRISPR genome editing utilizes Cas9 nuclease and single guide RNA (sgRNA), which directs the nuclease to a specific site in the genome and makes a double-stranded break (DSB). Design of sgRNA for CRISPR-Cas targeting, and to promote CRISPR adaptation, uses a regulatory mechanism that ensures maximum CRISPR-Cas9 system functions when a bacterial population is at highest risk of phage infection. Acinetobacter baumannii is the most regularly identified gram-negative bacterium infecting patients. Recent reports have demonstrated that the extent of diseases caused by A. baumannii is expanding and, in a few cases, now surpasses the quantity of infections caused by P. aeruginosa. Most Acinetobacter strains possess biofilm-forming ability, which plays a major role in virulence and drug resistance. Biofilm bacteria use quorum sensing, a cell-to-cell communication process, to activate gene expression. Many genes are involved in biofilm formation and the mechanism to disrupt the biofilm network is still not clearly understood. In this study, we performed in silico gene editing to exploit the AbaI gene, responsible for biofilm formation. The study explored different tools available for genome editing to create gene knockouts, selecting the A. baumannii AbaI gene as a target.