Ultra-conserved non-coding sequences within the FOXF1 enhancer are critical for human lung development


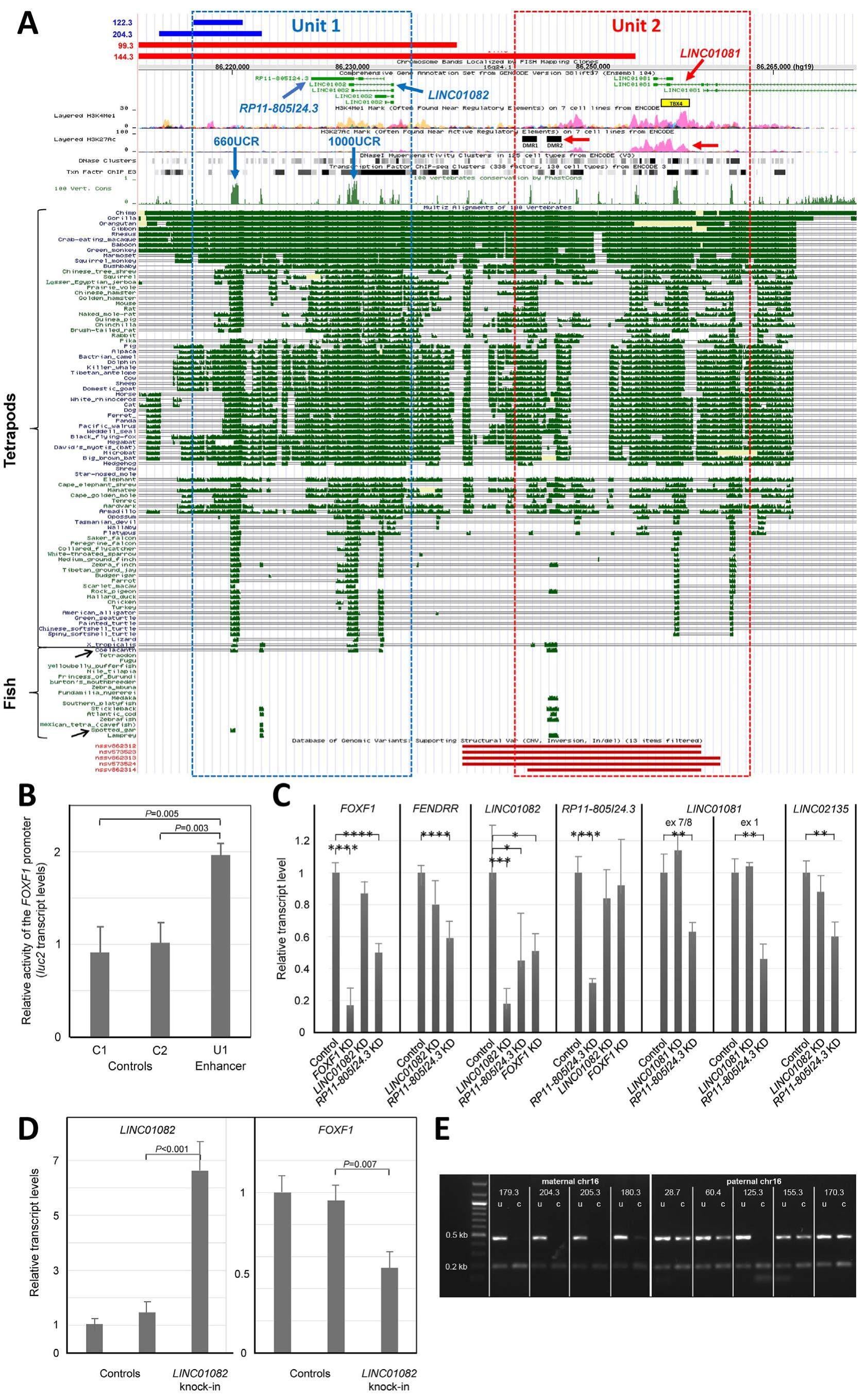
Heterozygous single nucleotide variants (SNVs) or copynumber variant (CNV) deletions, involving the mesenchymal forkhead box family transcription factor gene, FOXF1, or its distant lung-specific enhancer, are responsible for 80%-90% of cases of alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV). ACDMPV is a lethal lung developmental disorder with severe progressive respiratory failure and persistent pulmonary arterial hypertension (Supplemental Material). Intriguingly, in contrast to point mutations in FOXF1, the ACDMPV-causative CNV deletions arise de novo almost exclusively on the maternal chromosome 16q24.1. Thus far, we and others have described 50 de novo CNV deletions that arose on maternal chromosome 16 and only three de novo CNV deletions that arose on paternal chromosome 16q24.1. Here, we define an~660 bp ultra-conserved non-coding interval (660UCR) within the FOXF1 enhancer as critical for FOXF1 cis-regulation and normal human lung development. Heterozygous loss of this region on paternal chromosome 16 was found causative for ACDMPV. We also describe a novel enhancer lncRNA gene, RP11-805I24.3, located in the proximity to 660UCR and overlapping another ~1 kb ultra-conserved interval (1000UCR), as essential for the FOXF1 expression. Based on the obtained data, we propose a bimodal structure and parental functional dimorphism of the FOXF1 enhancer.