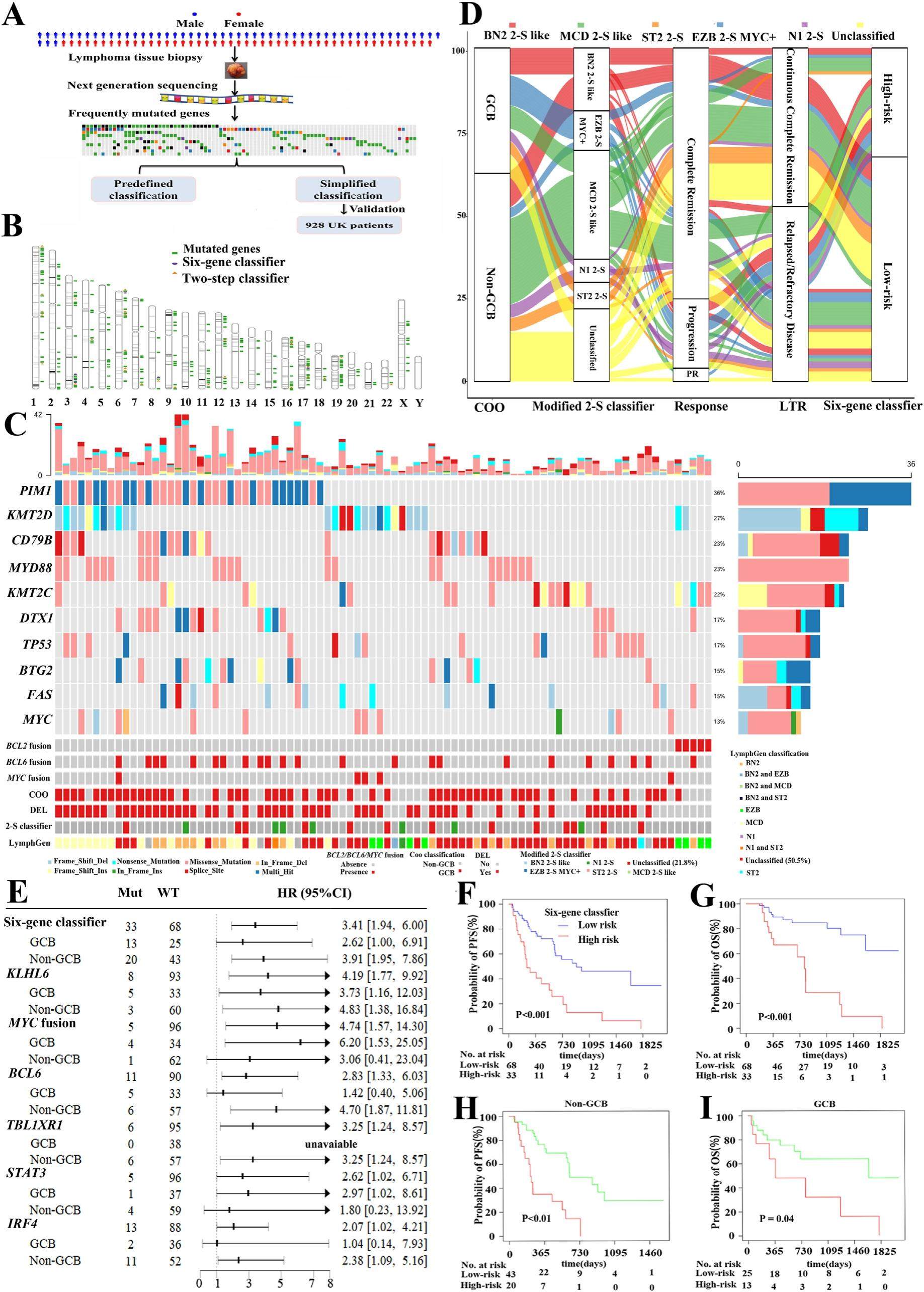
Simplifying genetic classifiers by six mutated genes in diffuse large B-cell lymphoma


Although diffuse large B-cell lymphoma (DLBCL) is considered as a curable disease after standard immunochemotherapy, approximately 30% of patients would succumb to short survival. To date, clinical presentations and gene expressions have been used to identify these high-risk patients. Recently, genetic alterations are used to model their proposed classifiers. For example, four, five and seven genetic subtypes were respectively found by Schmitz, Chapuy and George colleagues as a method for tailored treatment. However, these classifiers were calculated using the complex tools such as the LymphGen classifier and consensus clustering algorithm. Besides, mutated genes were not enough to estimate their genetic subtypes by their proposed tools, which need additional data such as copy number alteration and chromosome translocations. What's more, there is still not a consistent classifier to validate predefined subtypes. Thus, it is difficult to apply these genetic tools in clinical practice. In order to translate into clinical practice easily, a smaller gene-set using the easyto-implement method will be practical instead of measuring genome-wide sequencing. In this context, we searched for a small number of genes using targeted next-generation sequencing, and established its predictive ability for DLBCL patients.