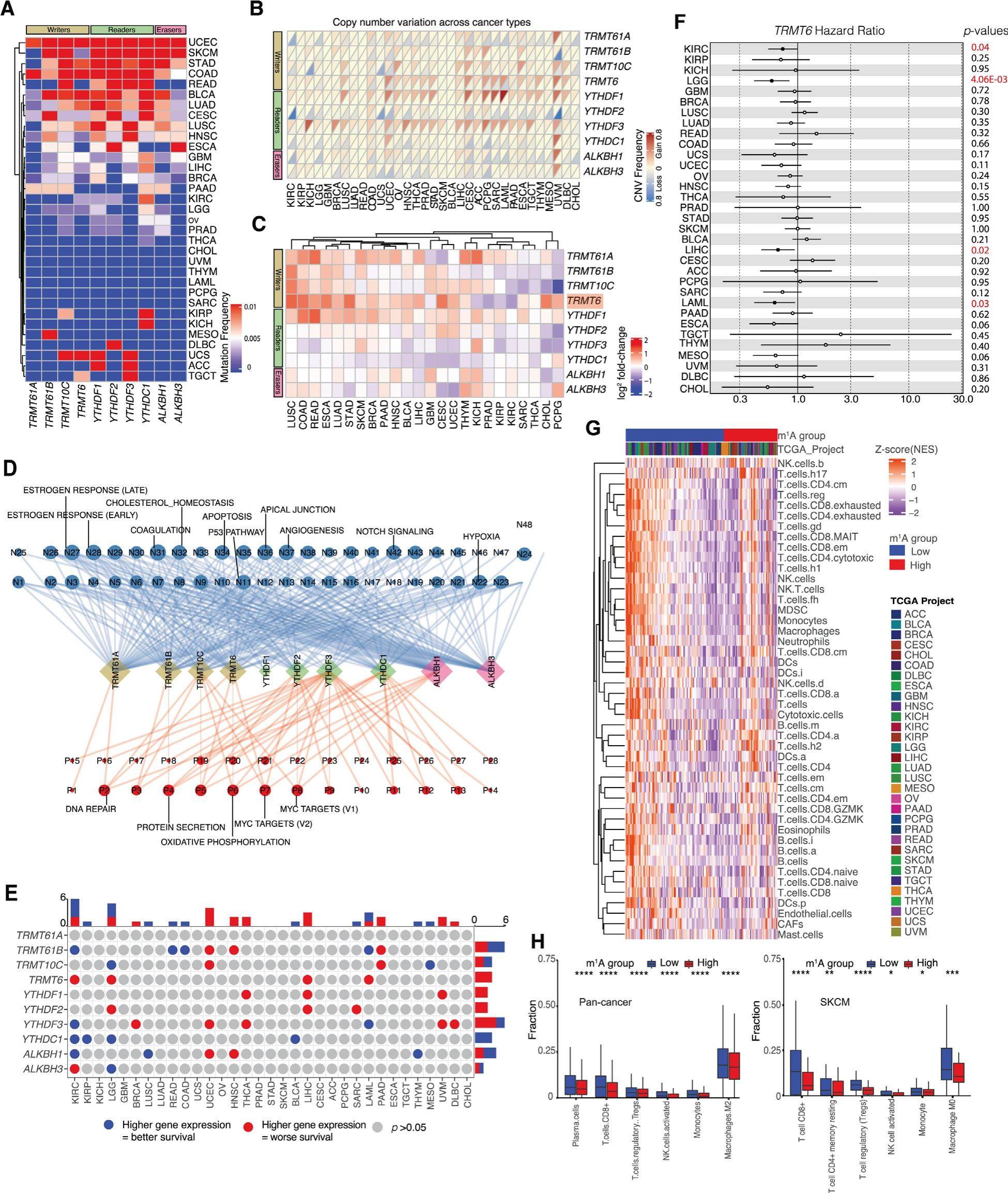
Altered expression of m1A regulatory genes is associated with oncogenic pathways, overall survival, and infiltration of immune cells in diverse human cancers


Of over 170 types of RNA modifications discovered, N6-methyladenosine (m6A) is the most actively studied. m6A confers oncogenic potential, maintains cancer stem cells, and is associated with cancer-related outcomes. The role of RNA modifications including m1A is poorly understood. m1A is abundant in transfer and ribosomal RNAs but has also been found on messenger RNAs. m1A stabilizes these RNA species to regulate essential biological processes including translation and ribosome biogenesis, which are frequently dysregulated in cancers. However, the role of aberrant m1A RNA methylation in cancer development and its utility in predicting clinical outcomes is currently unclear. Understanding the function of m1A RNA methylation in diverse cancers requires the characterization of genetic alterations and aberrant expression of m1A regulatory genes. These genes include m1A writers (TRM61A, TRIM61B, TRIMT61B, TRMT10C), erasers (ALKBH1, ALKBH3) and readers (YTHDF1, YTHDF2, YTHDF3 and YTHDC1). Here, we present the frequency of molecular abnormalities affecting m1A regulatory genes and their association with cancer-related pathways, prognosis, and tumor immune cell infiltration across 33 types of human cancers.