ScMicrobesAtlas: A comprehensive microbial atlas at single-cell resolution in human disease contexts


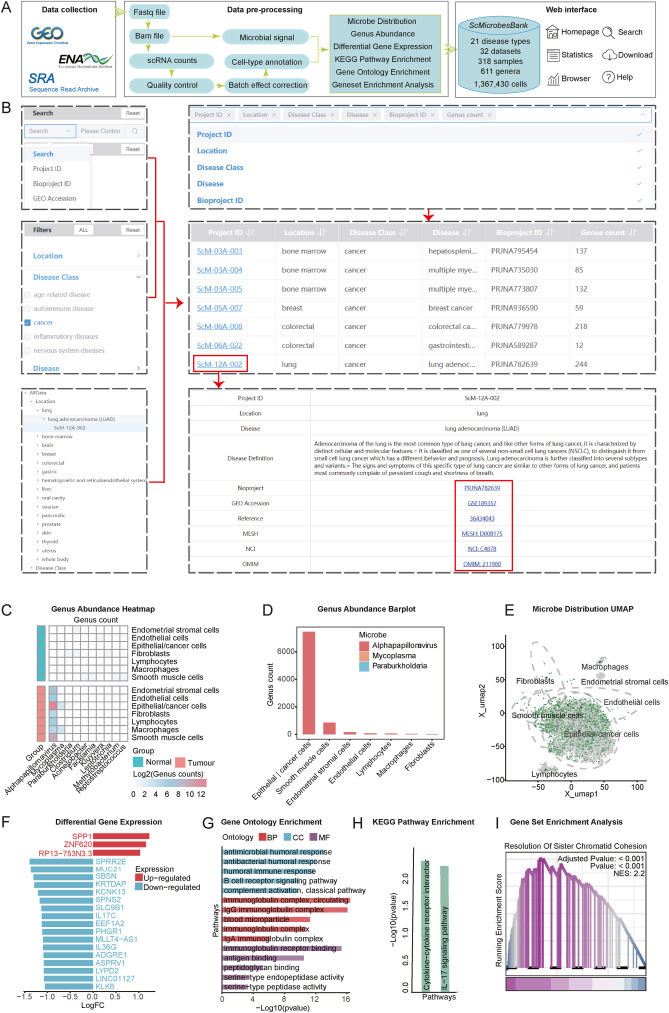
The microbiome plays a significant role in human health and disease.1 Although bulk tissue analyses have identified disease-specific microbial signatures, these studies do not capture microbial-host cell enrichments and their associations with cell-type-specific activities.2, 3, 4 Thus, we present the Single Cell Microbes Atlas (ScMicrobesAtlas, http://scmbdb.geneis.org.cn:8089), a comprehensive microbial atlas that provides single-cell resolution insights into various human diseases. To date, ScMicrobesAtlas version 1.0 integrates 318 samples from 32 single-cell RNA sequencing datasets, encompassing 21 disease types, and uncovers interactions between 611 bacterial genera and over 1.3 million human cells. All the data were uniformly processed with a standardized workflow, including quality control, batch effect removal, clustering, cell-type annotation, microbial signal identification and quantification, differential expression analysis, and functional enrichment analysis (Fig. 1A). ScMicrobesAtlas facilitates the comparative analysis of microbiome composition and identifies both shared and cell-type-specific microbial enrichments across diverse cell types and disease states. Additionally, it allows users to assess gene expression alterations and pathway activities in specific cell types in response to a particular microbiome. In summary, ScMicrobesAtlas offers a valuable resource for researchers to investigate disease-microbiome associations at the single-cell resolution.