A multi-omic integrative approach combining m6A-epitranscriptomic, transcriptomic, and splicing alternative events reveals potential candidates for colorectal cancer diagnosis


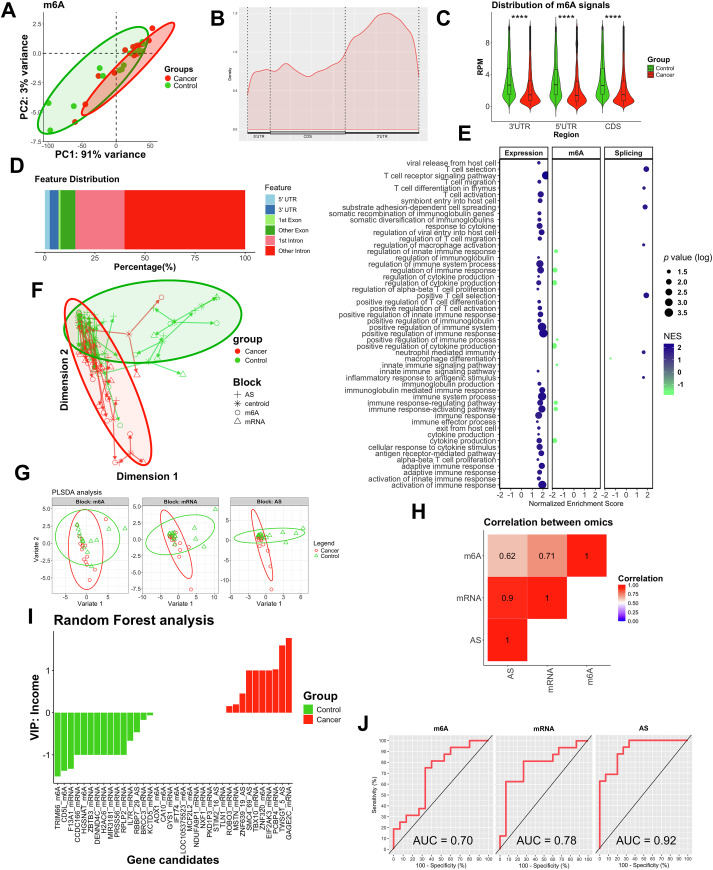
Colorectal cancer (CRC) continues to be the third most frequently diagnosed cancer, and the second leading cause of cancer-related mortality. Several non-invasive biomarkers have emerged, but only a few have been incorporated into clinical practice due to the lack of sensitivity.1 Research on the epigenome has unveiled potential clinical applications for diagnosis and therapy response.2,3 Particularly, recent evidence suggests a novel role of RNA methylation in the development of CRC,4 revealing an overall RNA m6A hypomethylation.5 However, our understanding of their contribution to CRC remains limited. To address this, we investigated m6A modification in CRC using an integrative approach. High-throughput sequencing was performed to analyze the m6A-epitranscriptome (methylated RNA immunoprecipitation sequencing; m6A), transcriptome (mRNA), and alternative splicing events (AS; RNA sequencing) in leukocytes from both healthy participants (n = 16) and patients with CRC (n = 15) (Table S1 summarizes the baseline characteristics of the participants) from the “Virgen de la Victoria” University Hospital, Málaga, Spain.