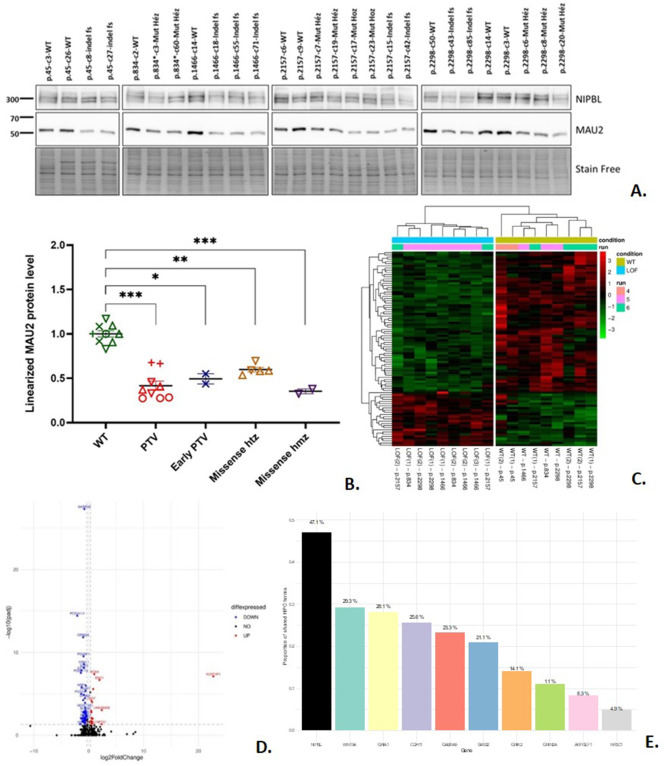
Assessment of the transcriptomic consequences and MAU2 protein levels in edited induced pluripotent stem cells with NIPBL pathogenic variants


Cornelia de Lange Syndrome (CdLS) is an intellectual disability syndrome characterized by distinctive clinical features including growth retardation, limb malformation, and a characteristic facial dysmorphism.1 Six genes, including NIPBL and MAU2, are associated with CdLS, all encoding components or partners of the cohesin protein complex. Cohesins play a central role in gene expression regulation by organizing chromatin and modulating transcription.2 CdLS is classified as a transcriptomopathy due to dysregulated transcription resulting from pathogenic variants in cohesin-related genes. NIPBL mutations are the most common cause of CdLS, impairing cohesin loading onto DNA. Previous transcriptomic studies have identified deregulated genes in CdLS (e.g., Liu et al3 or Mills et al4), but none have comprehensively assessed the impact of various NIPBL variants, including missense mutations, on isogenic induced pluripotent stem cells (iPSCs). In this study, we investigated mRNA and protein levels of NIPBL and MAU2, along with transcriptomic consequences of multiple NIPBL pathogenic variants, in isogenic iPSCs. Our findings confirm the role of NIPBL and MAU2 in CdLS pathogenesis and highlight deregulated genes contributing to the syndrome's phenotype.