Phosphorylation-related genes in lupus nephritis: Single-cell and machine learning insights


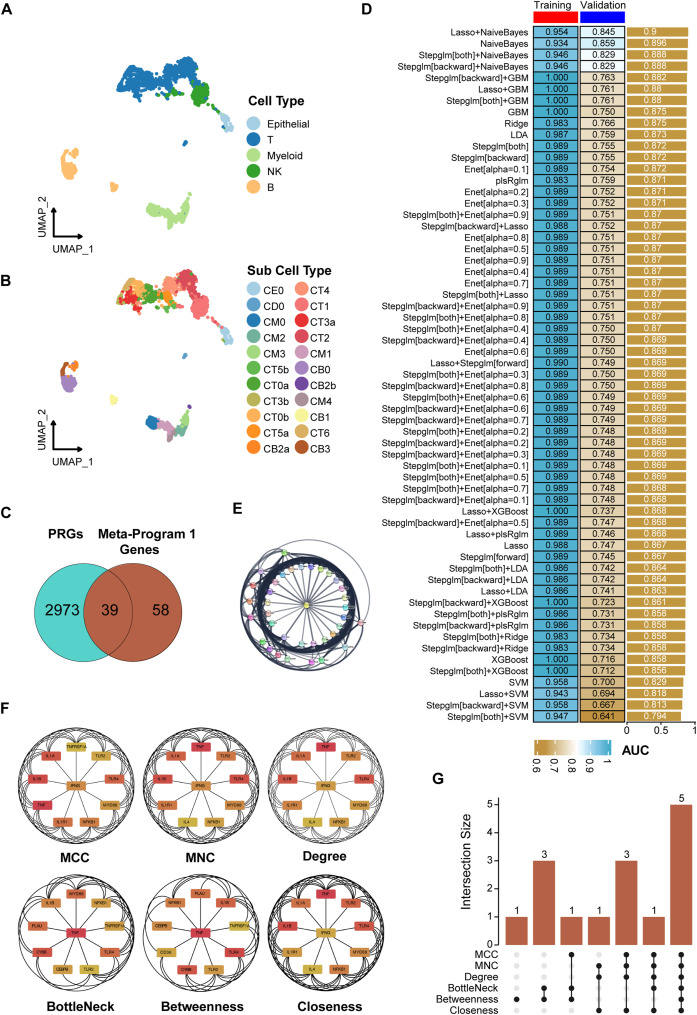
This study investigates key genes contributing to lupus nephritis (LN). While extensive research has elucidated various aspects of LN pathogenesis, the specific involvement of phosphorylation-related genes (PRGs) in this context remains an area of growing interest. We employ single-cell RNA sequencing analysis on renal tissues from 24 LN patients and 10 healthy controls. Leveraging the non-negative matrix factorization (NMF) algorithm, we identified critical gene patterns and constructed 61 predictive models using a comprehensive suite of 12 machine learning algorithms. We developed a predictive model using 6 PRGs, enhanced by a LASSO plus Naive Bayes approach. These PRGs demonstrated high predictive accuracy (the area under a receiver operating characteristic curve was 0.954 and 0.845 in training and validation cohorts, respectively). One of these PRGs, CEBPB (CCAAT enhancer binding protein beta), showed significant differences in expression between LN patients and controls. Further analysis revealed correlations between CEBPB and renal function parameters. The insights of this study into gene expression patterns enhance our understanding of the molecular mechanisms and support the potential for early diagnosis and targeted treatment strategies in LN. Extensive protein–protein interaction network analysis and clinical correlations underscore the important roles of these genes in LN. Our findings underscore the potential of PRGs as biomarkers, propelling the precision medicine frontier in LN diagnostics and therapeutics.