Common interacting genetic variation shapes susceptibility to type 1 diabetes in a Colombian Caribbean community: In search of shared genetic markers


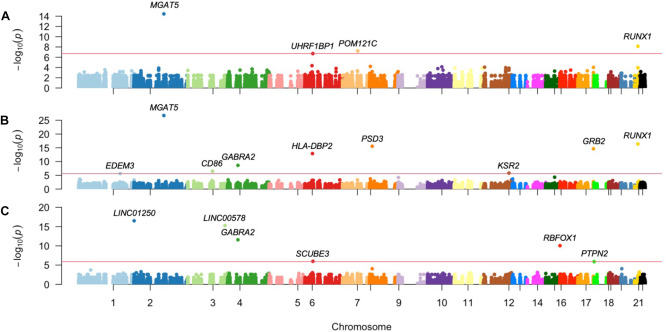
Genome-wide association studies (GWASs) have identified hundreds of loci across the human genome conferring susceptibility to autoimmune diseases (AIDs), some of which are shared between more than two diseases. However, this univariate approach has limitations in detecting complex genotype-phenotype correlations. In this work, we carried out whole-exome sequencing of Colombian Caribbean patients with type 1 diabetes (T1D), lupus nephritis (LN), and juvenile idiopathic arthritis (JIA), to evaluate functional exomic variation, i.e., single nucleotide polymorphisms (SNPs), and to outline common and rare variations underpinning the susceptibility to these autoimmune diseases. Single and multi-locus linear mixed-effects models fit the data to identify T1D-associated genomic variants and the most likely genetic architecture underpinning AID risk. Variations associated with T1D susceptibility pointed to genes related to glycoprotein oligosaccharide biosynthesis, phospholipid binding, pancreatic adenocarcinoma, systolic blood pressure, and fasting insulin metabolism, among others that highlight MGAT5 (PFDR = 1.64 × 10−22), RUNX1 (PFDR = 1.8 × 10−12), PSD3 (PFDR = 8.1 × 10−12), and HLA-DBP2 (PFDR = 2.18 × 10−9). Our study outlines oligogenic common variation underpinning the susceptibility to develop T1D. These genetic polymorphisms are also shared by patients with other AIDs such as LN and JIA, indicating that the shared genetic architecture (defined by pleiotropy and epistasis) shapes the genetic susceptibility of these disorders in this multiethnic population.