Single-cell transcriptome profiling reveals altered neural crest cell dynamics and novel biomarkers in EDNRB mutant mice with Hirschsprung's disease phenotype


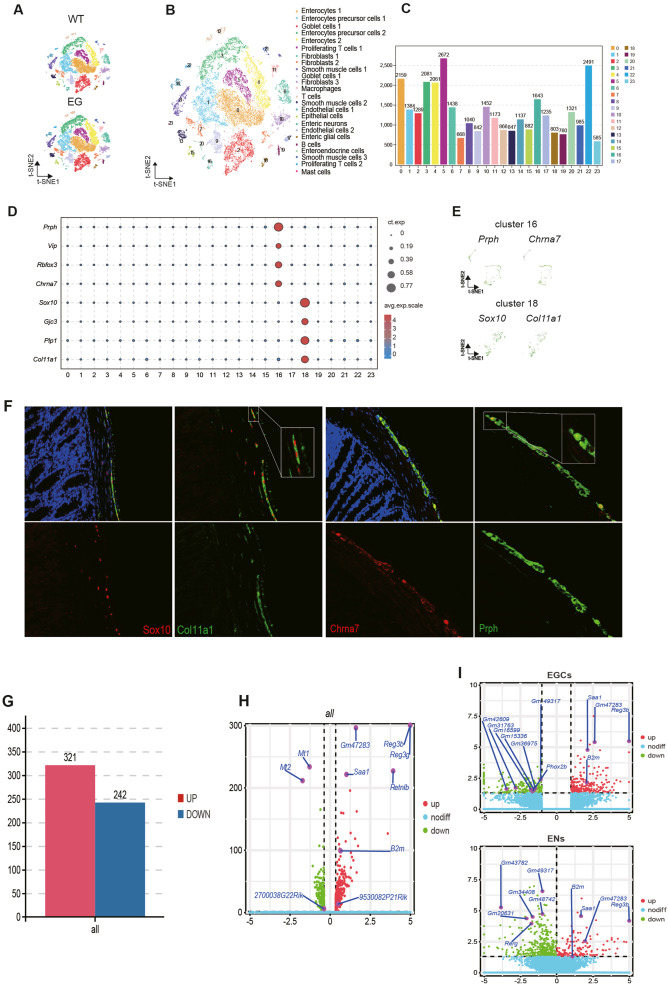
Hirschsprung's disease (HSCR) results from neural crest cell migration and differentiation issues.1 Previously, analysis was limited to bulk tissue samples, but single-cell RNA sequencing now offers microscopic insights. Our pioneering single-cell RNA sequencing analysis of colon tissue is helping unravel HSCR's origins and development. We examined 22,353 cells from wild-type (WT) and endothelin receptor type B (EDNRB) mutant mice, identifying 24 clusters comprising 15 cell types. Comparing HSCR-type and healthy colons revealed distinctions in cellular differentiation, transcriptome characteristics, and biological functions. Our goal is to fill knowledge gaps and uncover the molecular basis of HSCR, facilitating potential therapeutic advancements.