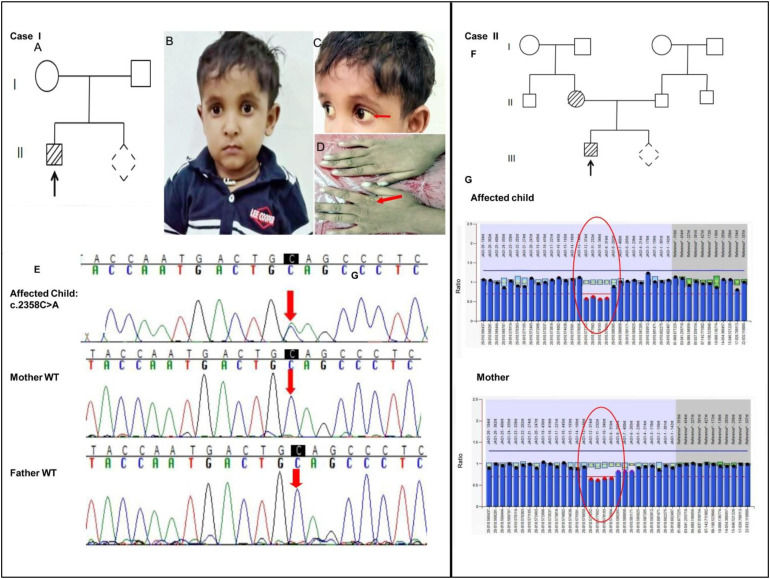
Two unrelated Alagille syndrome cases of South Indian origin: Showing multi-exonic deletion and a novel mutation in JAG1 gene


Alagille syndrome (ALGS; OMIM #118450) is a highly variable, multi-systemic, autosomal dominant disorder, caused by mutations in the Jagged Canonical Notch Ligand 1 (JAG1) gene (20p12.2) or in the Neurogenic locus notch homolog protein 2 (NOTCH2) gene (1p13).1 It primarily affects the liver, heart, eyes, face, kidney, skin, vertebrae, and skeleton.1,2 The JAG1 gene has 26 exons and about 90% of the ALGS cases result from pathogenic variants in the JAG1 and around 7% of cases have deletions in chromosome 20 that include this gene.1,3 Latest reports reveal that the spectrum of mutations includes 75% protein-truncating mutations and the rest 25% are non-protein-truncating mutations. Few cases (<1%) have also been reported with pathogenic variants in a second gene called NOTCH 2 gene.1,3,4 Various studies have elucidated that ALGS clinical phenotype is caused by different pathogenic mutations in JAG1 and NOTCH2 genes suggesting the haploinsufficiency of these two genes as the primary mechanism for disease pathobiology rather than a dominant negative mechanism.1,2 Here we present a case report of two protein-truncating JAG1 mutations detected in two unrelated cases of South Indian origin.