Novel virulence-related genes that contribute to clinical infections of Salmonella enteritidis


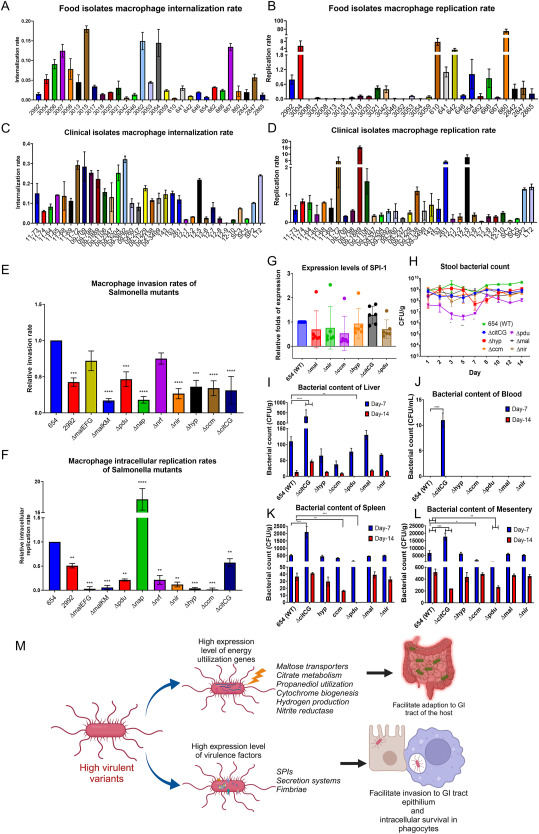
Salmonella is a notorious foodborne pathogen that comprises strains that exhibit varied ability to cause human infection. To date, this pathogen still causes over one million cases of foodborne infections annually in the United States alone.1 The systemic infections of high-virulence Salmonella strains are often seen in the nosocomial environment. The increased prevalence of antimicrobial-resistant genes in highly virulent Salmonella causes their invasive infection more difficult to treat. Therefore, understanding the mechanism of Salmonella virulence is important for solving public health issues. According to the information from the US CDC, Salmonella enteritidis (S. enteritidis) infection is the most common cause of Salmonella infection in clinical cases among different serotypes.1 We collected a set of high- and low-virulence S. enteritidis isolates and subjected them to comparative genomic, transcriptomic, and phenotypic analyses. The tested strains exhibited almost identical genetic composition, but over-expression of genes involved in various physiological functions was observed in the high-virulence strains. Importantly, these genes include those responsible for maltose transport, citrate metabolism, VitB12 biosynthesis, propanediol utilization, nitrite reduction, and hydrogen production. The gene knockout experiment confirmed that the deletion of these genes resulted in decreased invasiveness, reduced survival inside macrophages, reduced invasion of different organs, and lower mortality in animal experiments.