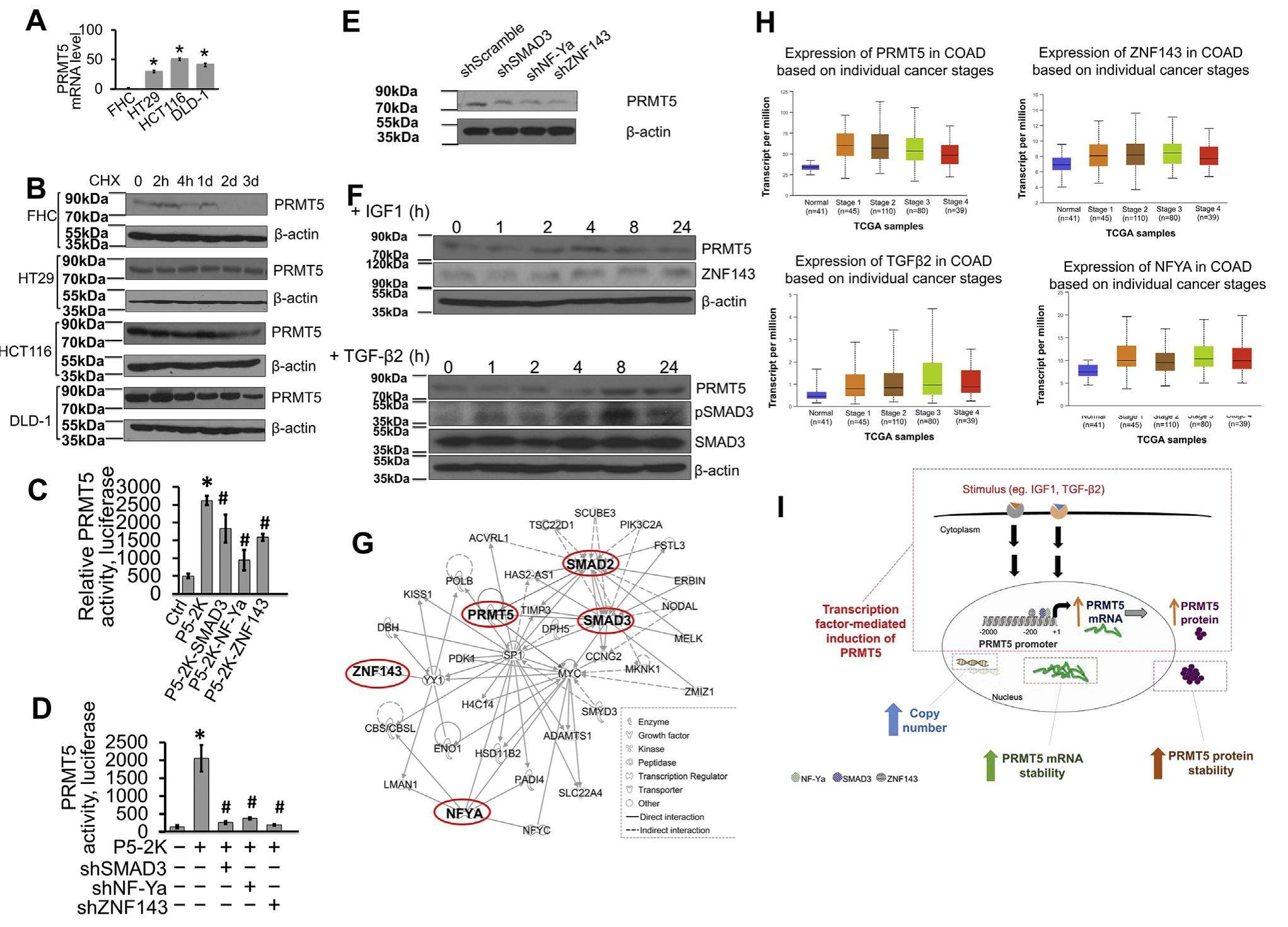
A complex signature network that controls the upregulation of PRMT5 in colorectal cancer


Protein arginine methyltransferase 5 (PRMT5) deregulation has emerged as an important prognostic indicator in human cancers. Through aberrant methylation-mediated epigenetic modification of signaling molecules, PRMT5 overexpression contributes to dysregulation of a variety of cellular processes related to cancer progression. However, themechanismsgoverningPRMT5expressionlevelsincancer remain largely unknown. In this study, we examined factors that regulate PRMT5 expression at multiple levels. We mapped three regions of the proximal promoter of PRMT5 and identified NF-Ya, SMAD3, and ZNF143 as part of a key signature network node regulating PRMT5 expression in HT29 colorectal cancer (CRC) cells. Importantly, we provide evidence that knockdown or ligand induced activation of SMAD3, and ZNF143 led to changes in PRMT5 transcript and protein levels, respectively. We showed that PRMT5 expression positively correlates with both TGF-β2 and ZNF143 expression, suggesting that activation and/or upregulation of these proteins may be partly responsible for PRMT5 overexpression in a subset of CRC patients. Collectively, our data present a complex model that involves cell-autonomous induction of PRMT5 in CRC cells by transcriptional mechanisms and upregulation of PRMT5 mRNA and protein, which encompasses processes involving gene amplification and increased transcription and protein turnover rates.