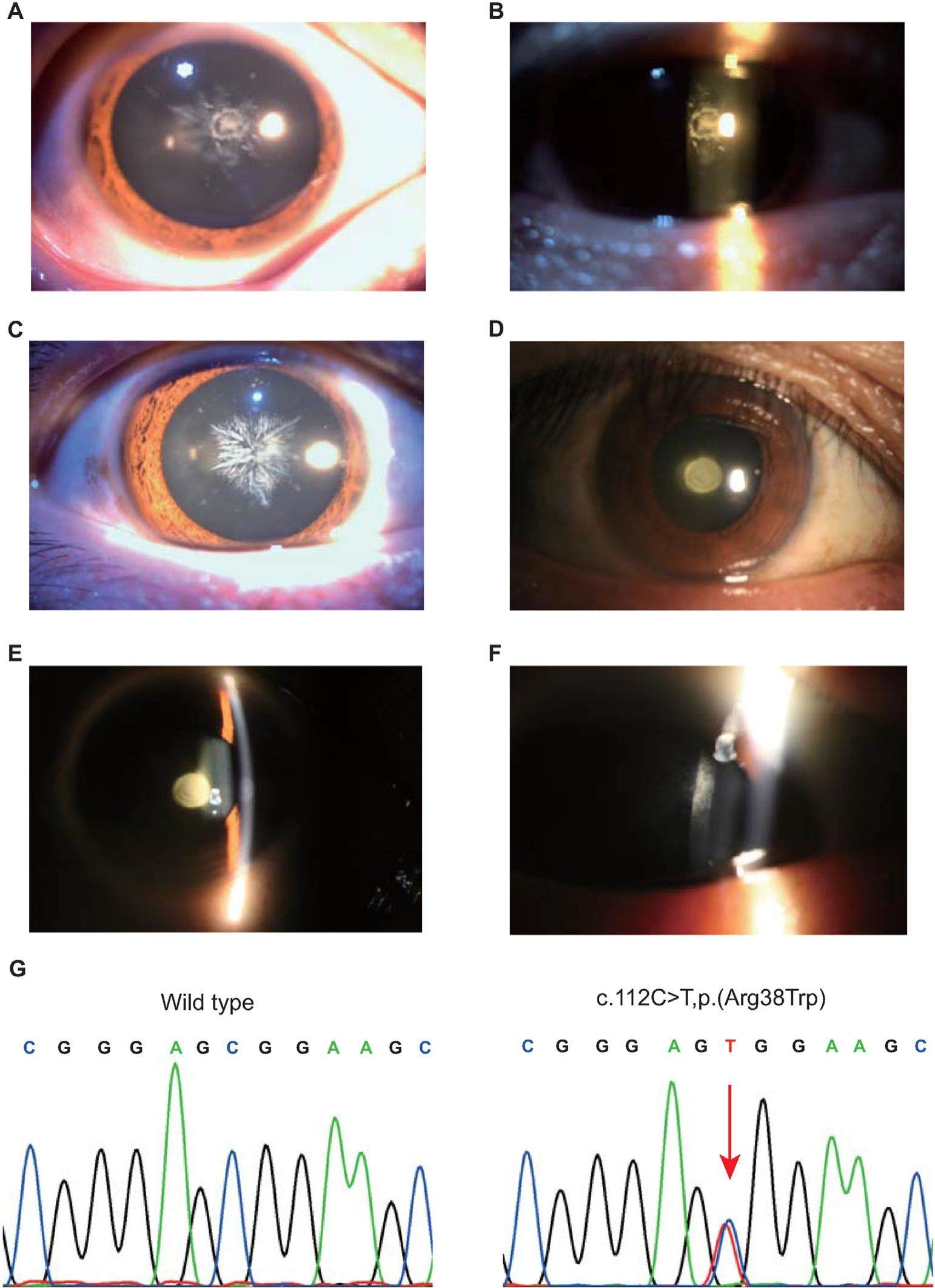
Variations in EXD3 caused congenital cataracts in three Chinese families


Congenital cataract is a clinically and genetically heterogeneous disease characterized by any opacity of the lens presenting at birth or in the first year of life, with an incidence of 1-6/10,000 live births in developed countries and 5-15/10,000 live births in developing countries. Congenital cataract accounts for 7.4%-15.5% of all childhood blindness, and it occurs as either a syndromic disease or an isolated (nonsyndromic) disease with or without other ocular malformations, such as microcornea, microphthalmia, persistent fetal vascularization, glaucoma, or retinal dystrophies. Whereas autosomal dominant inheritance is most common, autosomal recessive and X-linked inheritance have also been reported, indicating a degree of genetic heterogeneity in congenital cataract. To date, various cataract-associated loci have been detected, with over 453 genes associated with cataracts reported prior to October 1,2021 (http://cat-map.wustl.edu/). The pathogenic variation detection rate in familial cases was approximately 75%, indicating that the genetic basis for approximately one-fourth of familial patients is still unknown. In recent years, there has been a shift from traditional genetictests (e.g., Sanger sequencing) to genomic tests (comparative genomic hybridization (CGH)), gene panels, and next-generation sequencing (NGS) technologies) for many diseases with complex inheritance and genotypes, especially rare Mendelian diseases. NGS technologies, such as whole-exome sequencing (WES) and wholegenome sequencing (WGS), are important for identifying pathogenic variations, especially for discovering new pathogenic genes in Mendelian diseases with genetic heterogeneity. With the development of NGS technologies, more disease-causing genes associated with congenital cataract will be discovered. In this study, we identified a novel association between a missense variation in EXD3 and congenital cataract in three Chinese families by combining NGS technologies and linkage analysis.