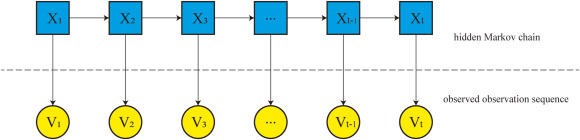
The hidden Markov model and its applications in bioinformatics analysis


Big biological data contains a large amount of life science information, yet extracting meaningful insights from this data remains a complex challenge. The hidden Markov model (HMM), a statistical model widely utilized in machine learning, has proven effective in addressing various problems in bioinformatics. Despite its broad applicability, a more detailed and comprehensive discussion is needed regarding the specific ways in which HMMs are employed in this field. This review provides an overview of the HMM, including its fundamental concepts, the three canonical problems associated with it, and the relevant algorithms used for their resolution. The discussion emphasizes the model's significant applications in bioinformatics, particularly in areas such as transmembrane protein prediction, gene discovery, sequence alignment, CpG island detection, and copy number variation analysis. Finally, the strengths and limitations of the HMM are discussed, and its prospects in bioinformatics are predicted. HMMs can play a pivotal role in addressing complex biological problems and advancing our understanding of biological sequences and systems. This review can provide bioinformatics researchers with comprehensive information on HMM and guide their work.