Mutational and transcriptional profiling of cuproptosis-associated genes in amyotrophic lateral sclerosis


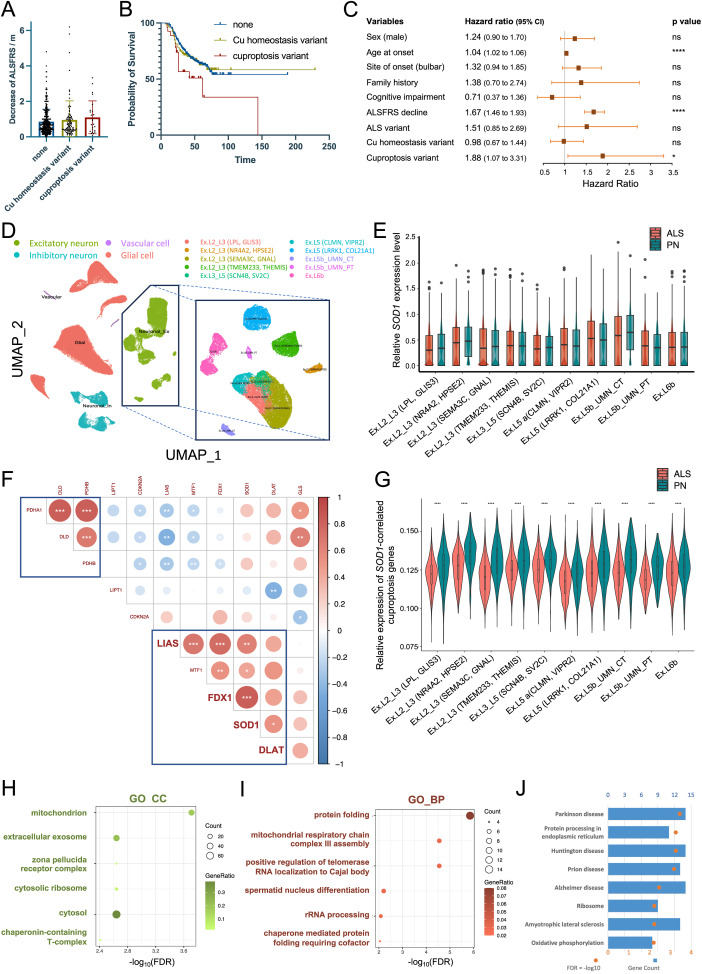
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder characterized by progressive muscle weakness and atrophy.1 The widespread implementation of next-generation sequencing in clinical practice has facilitated the identification of over 40 ALS-associated genes, among which copper/zinc-superoxide dismutase 1 (SOD1) assumes a pivotal role in East Asian populations.1 Moreover, it is increasingly apparent that the intricate interplay between genetic predisposition and exposome over time may contribute significantly to the etiology of this debilitating disease. While the precise pathogenic role of copper (Cu) in ALS remains elusive, accumulating evidence suggests that Cu deficiency may be linked to SOD1-associated ALS, possibly mediated through the promotion of the hydrophobicity of mutant SOD1, ultimately exacerbating its neurotoxic aggregation.2 Furthermore, a novel form of regulated cell death, termed cuproptosis, has emerged as an intriguing phenomenon. Cuproptosis is characterized by the direct binding of Cu to the lipoylated enzymes of the tricarboxylic acid cycle, leading to mitochondrial lipoylated protein aggregation and consequent proteotoxic stress.3 Given that mitochondrial dysfunction constitutes a critical pathophysiological hallmark in ALS, the perturbation of Cu homeostasis may possibly play a prominent role in the context of SOD1-induced neurodegeneration.