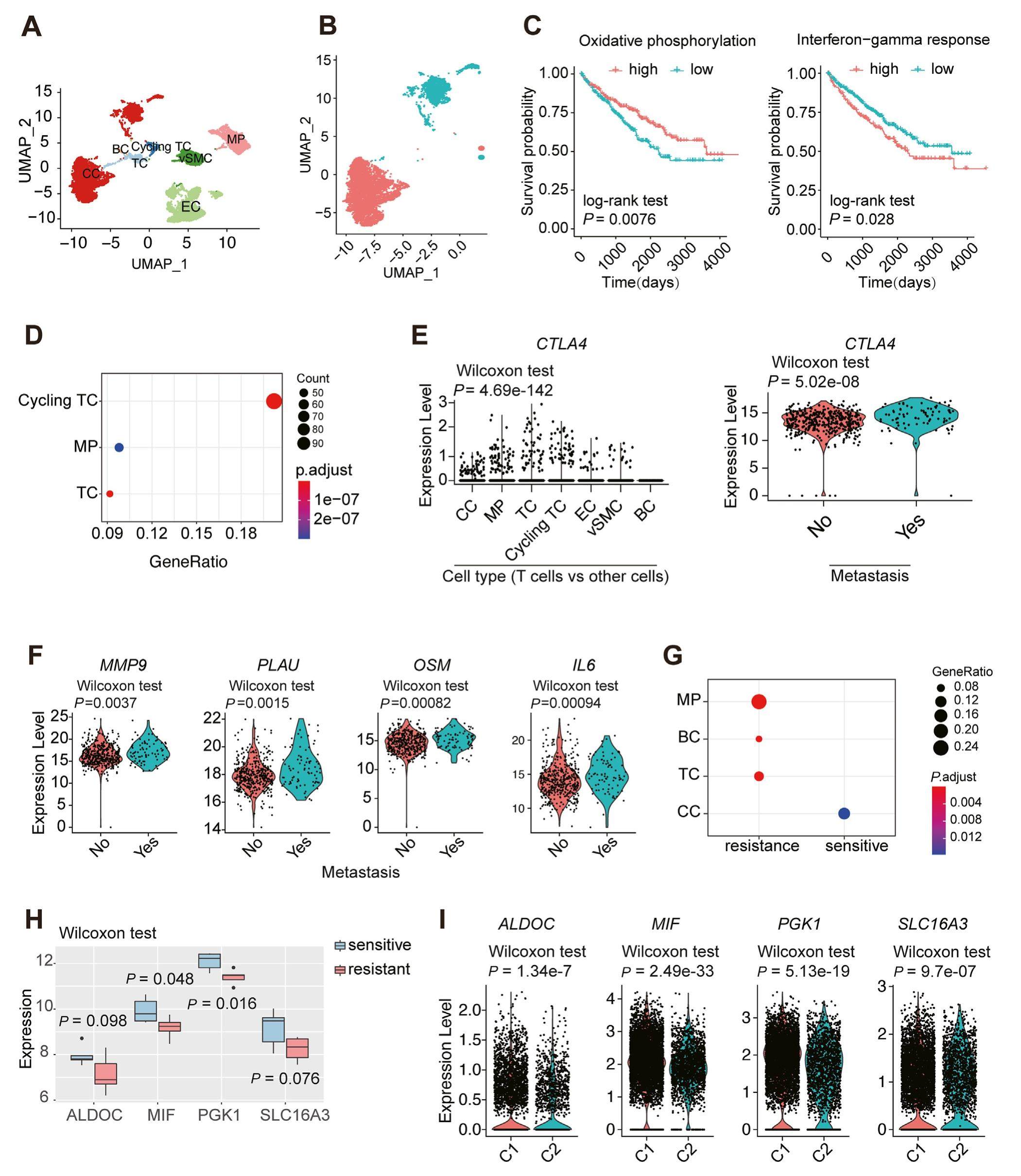
Integrated bulk and single-cell transcriptome data identify clinically relevant cell populations in clear cell renal cell carcinoma


Renal cell carcinoma, which consists of three main histological subtypes, namely, clear cell renal cell carcinoma (ccRCC), papillary RCC (pRCC), and chromophobe RCC (chRCC), accounts for most kidney cancer cases. ccRCC accounts for approximately 85% of all RCC cases, and it is a malignant tumor with multiple molecular features and a poor prognosis. Studies have shown that ccRCC is one of the most immune and vaso-invasive cancer types. Considering that ccRCC is not sensitive to either radiotherapy or chemotherapy, targeted therapies, such as vascular endothelial growth factor inhibitors/tyrosine kinase inhibitors and immunotherapy, are now the mainstay first-line therapies that are used in the clinic. However, a significant proportion of patients do not respond to the two treatments mentioned above or have high recurrence rates, which are related to the high degree of tumor heterogeneity and the tumor microenvironment (TME) of ccRCC. Single-cell transcriptome sequencing provides accurate gene expression data at the single-cell level. Integrated single-cell and large transcriptome data from ccRCC samples provide comprehensive information, such as information about prognosis, metastasis, and response to sorafenib and anti-PD-1 treatment. In the present study, we aimed to integrate bulk and single-cell transcriptome data to reveal the clinically relevant cell types in renal cell carcinoma, aiming to reveal the underlying molecular mechanism.