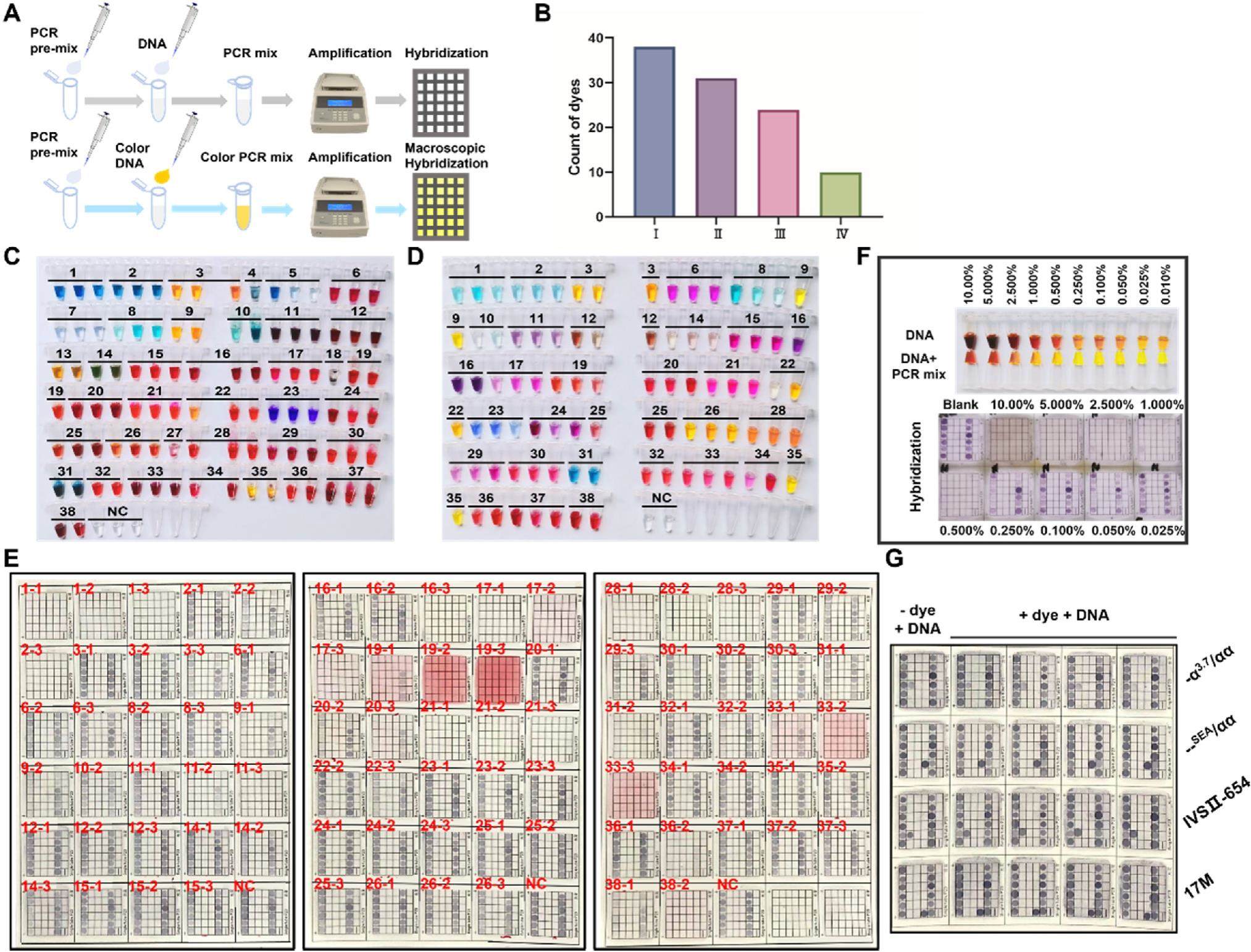
A color-based FHGC approach facilities DNA-based clinical molecular diagnostics


The flow-through hybridization and gene chip (FHGC) was developed to used in clinical molecular diagnostics for thalassemia, human papillomavirus (HPV), and other diseases. FHGC could improve hybridization efficiency and reduce hands-on time, thus improving precision, reproducibility, and traceability. Multiple genotypes can be detected simultaneously without incurring high costs. During the experiment, the polymerase chain reaction (PCR) product or buffer forced through the membrane matrix, PCR products are apprehended by a probe attached to the membrane, and molecular typing of the disease or microorganism was determined after subsequent amplification by the streptomycin avidin (the details are given in the Supplementary File). However, the steps prior to hybridization, such as DNA extraction, PCR amplification, and sample loading into the hybridization wells, require a significant amount of manual operation. Each batch involves handling dozens of samples, and Eppendorf (EP) tubes used to load the amplification mixture are typically packed tightly together. Furthermore, the reagent is colorless and transparent, making it difficult to determine whether the sample was loaded into EP tube or not with a very small sample size. Operator carelessness or distraction will result in errors and unnecessary trouble.